Projects
Checkout my GitHub for code of selected projects
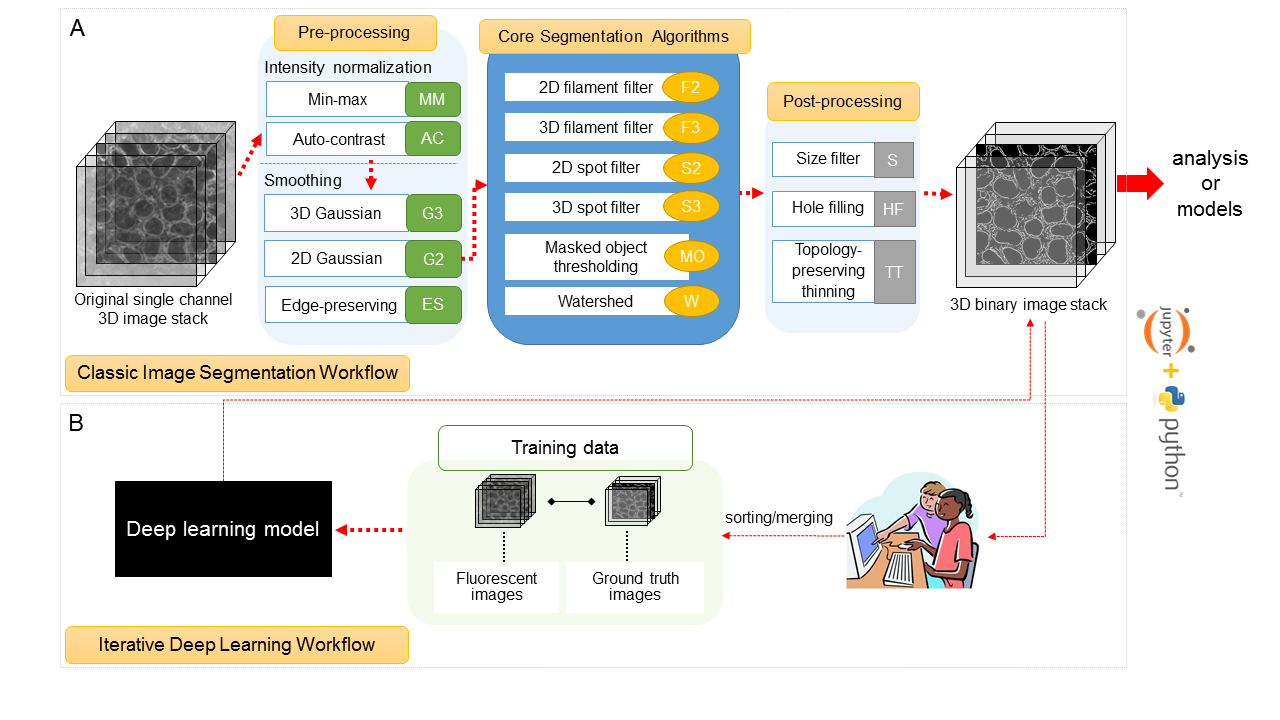
Allen Cell Structural Segmenter
-
Allen Institute for Cell Science
Python-based open source toolkit developed at the Allen Insttitue for Cell Science for 3D segmentation of intracellular structures in fluorescence microscopic image. - Allen Cell Structural Segmenter is powerful tool that can generate and curate training dataset, train deep learning model, and segment structures with trained model. Multiple modules and networks that I worked on are integrated into this open source toolkit. I developed and packaged deep learning-based cell pair detector by implementing Faster-RCNN. Furthermore, with spatial data augmentation and variant form of DeeplabV3+, I developed a deep learning based 2D segmentation tool for the highly complex shaped cell structures. This framework is very versatile and can be applied to various types of cell detection tasks. It is now available in AWS and Napari as plug-in.
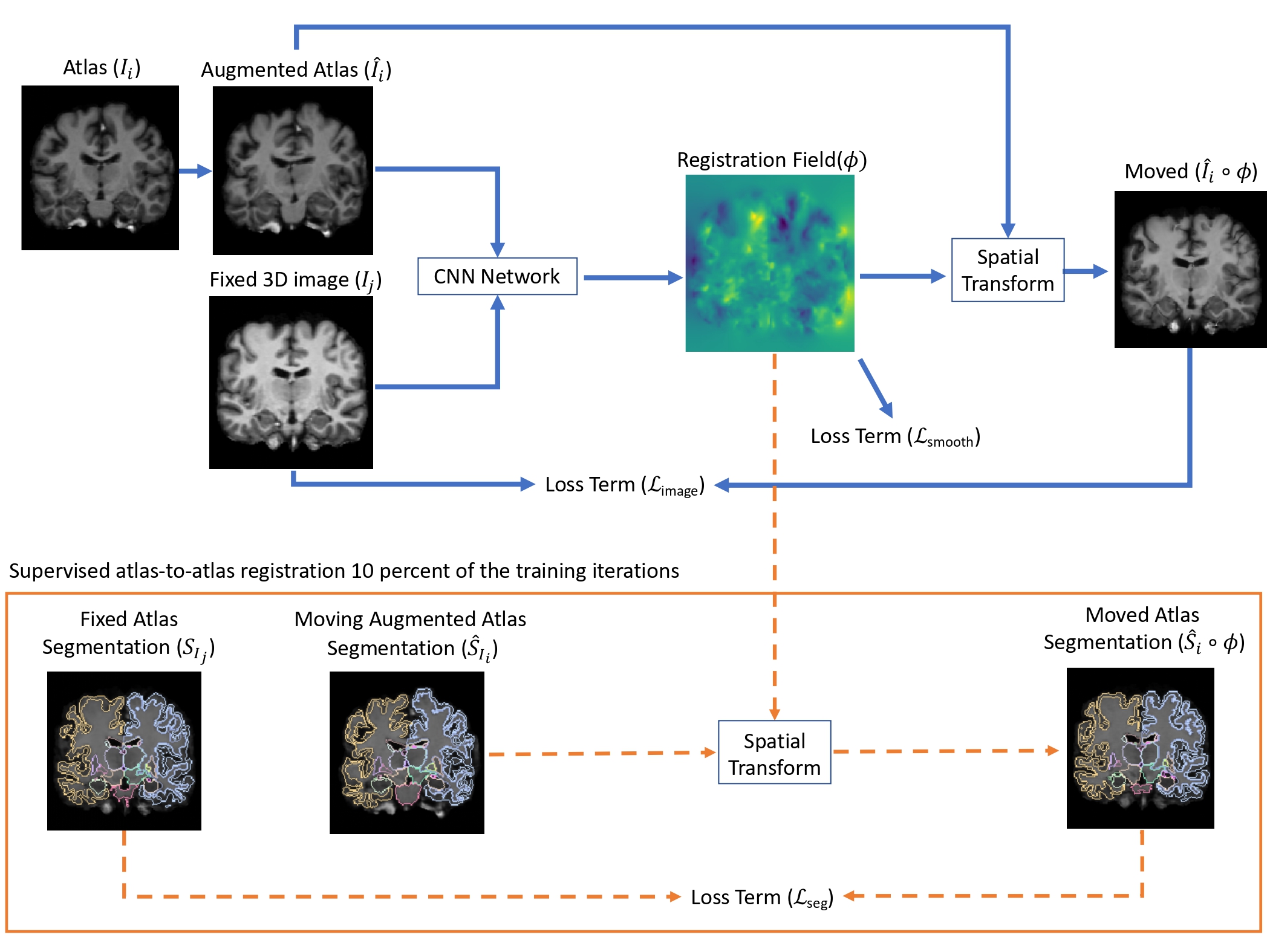
3D Brain Segmentation with Multi-Atlas Segmentation
-
Sabuncu Lab, Cornell University
Proposed and implemented 3D segmentaiton algorithm using multi-atlas segmentation with sermi-supervised registration with brain MR image dataset. - Supervised by Dr.Mert Sabuncu and Dr.Adrian Dalca. We tackled biomedical image segmentation in the scenario of only a few labeled brain MR images exist. We proposed a straightforward implementation of efficient semi-supervised learning-based registration method, which we showcase in a multi-atlas segmentation framework (Voxelmorph). Second, through an extensive empirical study, we evaluate the performance of a supervised segmentation approach, where the training images are augmented via random deformations. I have implemented supervised segmentation model with U-Net using Tensorflow framework (repo). Surprisingly, we find that in both paradigms, accurate segmentation is generally possible even in the context of few labeled images.